Abstract
The first two authors and last two authors contributed equally.
Genome-wide association studies (GWAS) have identified risk loci for Acute Lymphoblastic Leukemia (ALL), Chronic Lymphoblastic Leukemia (CLL) and Non-Hodgkin Lymphoma, however an Acute Myeloid Leukemia (AML) GWAS has not been published to date. We performed a GWAS to identify AML and Myelodysplastic Syndrome (MDS) risk loci using a nested case-control study design in the DISCOVeRY-BMT cohorts which includes almost 2000 AML and MDS patients as cases and 2813 unrelated donors as controls.
Genotyping was performed using the Illumina Human OmniExpress BeadChip and imputed using the Haplotype Reference Consortium, yielding > 8 million high-quality variants for statistical analysis. Logistic regression models with AML (de novo AML with normal cytogenetics, de novo AML with abnormal cytogenetics and therapy-related AML) and MDS cases and European American healthy donor controls adjusted for age and sex were used to test the association of each SNP with disease status. To identify the strongest association signal with disease we conducted a summary statistic SNP-based association analysis (ASSET) using non-overlapping AML and MDS cases implemented in R statistical software. ASSET uses an exhaustive search for SNPs with small but common pleiotropic effects across groups of traits while accounting for the multiple tests required by the subset search, as well as any shared controls between groups. This approach allowed us to further investigate the heterogeneity within AML subtypes and to gain increased power by pooling subtypes that show pleiotropic effects. ASSET genome wide (GW) significance is defined as P<5.0x10-8, however, we present results of each subset analysis.
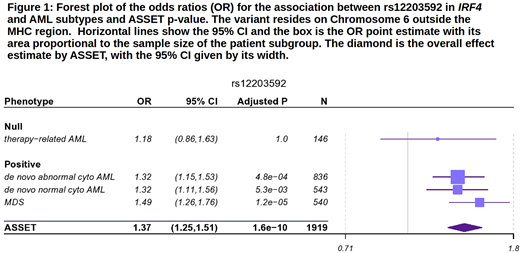
ASSET analyses identified an association of T allele at rs12203592, Interferon Regulatory Factor 4 (IRF4), with increased risk of de novo AML and MDS (Figure 1) (OR=1.37; 95% CI, 1.25-1.51, Pmeta=1.59x10-10). The variant has been reported significant in GWAS of skin pigmentation, hair color, non-melanoma skin cancer, squamous cell carcinoma, actinic keratosis, and childhood ALL; GW case-control studies of neuroblastoma and breast cancer show suggestive association signals (P<5 x 10-6).
IRF4 belongs to the IRF family of transcription factors and is a key regulator of differentiation stages in hematopoiesis. The T allele at rs12203592 is associated with significantly increased expression of IRF4 in whole blood and lung tissue and in vitro mouse studies have shown the T allele to be associated with higher levels of IRF4 transcription. rs12203592 is <80bp from an IRF4 transcription start site and in an important position within NF-κB motifs in multiple blood and immune cell lines, supporting the hypothesis that this SNP modulates NF-κB repression of IRF4 expression.
Another variant in the IRF4 regulatory region, rs62389423 (A allele), showed a putative association with subsets of de novo AML and MDS (OR=1.36; 95% CI,1.21-1.52, Pmeta=1.2x10-7). Although not in linkage disequilibrium with rs12203592, the A allele has also been previously associated with susceptibility to skin cancer and melanoma in multiple GWAS. Analysis of 596 UKbiobank CLL cases and >300,000 controls, shows the A allele correlates with a 50% increased risk of CLL (95% CI, 1.32-1.73, P= 2.4x10-9).Several other GWAS also show an association between additional IRF4 variants and CLL.
Analyses in individual subtypes also revealed an intronic variant, rs10098598, in RAD21 to be associated with de novo AML (OR=2.18; 95% CI, 1.60-2.97; Pmeta=8.9x10-7). RAD21, on8q24, a region known to be associated with multiple cancer types, is involved in DNA double-stranded break repair and in chromatid cohesion in mitotic cells. Loss of function would theoretically lead to chromosomal instability and tumorigenesis. Components of the cohesion complex (including RAD21) are somatically mutated in ~12% of de novo AML and MDS patients, where it is frequently an early event.
We provide the first GW evidence of association between a common variant and AML susceptibility. This SNP has been shown to be associated with multiple phenotypes, suggesting that there are pleiotropic effects at work. Our RAD21 finding is consistent with the role of cohesion in leukemogenesis and provides some evidence of its role in de novo AML susceptibility. Replication and further subset analyses with genome-wide data in >2000 AML cases are ongoing.
Griffiths:Alexion Inc.: Honoraria, Research Funding; Astex/Otsuka Pharmaceuticals: Honoraria, Research Funding; Celgene, Inc: Honoraria, Research Funding; Novartis, Inc.: Research Funding; Pfizer, Inc.: Research Funding. McCarthy:Bristol Myers Squibb: Consultancy, Honoraria; Celgene: Consultancy, Honoraria, Research Funding; Karyopharm: Consultancy, Honoraria; Takeda: Consultancy, Honoraria; Janssen: Consultancy, Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal